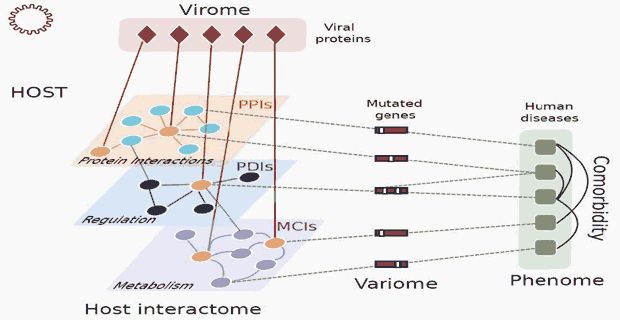
 Many virally implicated human diseases are associated with genetic alterations in particular disease susceptibility genes. For two human DNA tumor viruses, Epstein-Barr virus and human papillomavirus 16, we introduced a network-based framework to model the phenotypic consequences of the viral-host interactions (Gulbahce, Yan, et al, PLoS Comput Biol 2012). We found that
Many virally implicated human diseases are associated with genetic alterations in particular disease susceptibility genes. For two human DNA tumor viruses, Epstein-Barr virus and human papillomavirus 16, we introduced a network-based framework to model the phenotypic consequences of the viral-host interactions (Gulbahce, Yan, et al, PLoS Comput Biol 2012). We found that
- Viral targets of EBV and HPV16 proteomes are significantly closer to genes associated with virally implicated diseases than to genes associated with random diseases. Given this proximity, genes in the neighborhood of viral targets in the host interactome are more likely associated with virally implicated diseases, compared to genes elsewhere in the host interactome (local impact hypothesis)
- For representative EBV- and HPV16-implicated diseases, genes in the neighborhood of viral targets in the host interactome have significantly altered expression levels in virally implicated disease tissues, in line with the local impact hypothesis.
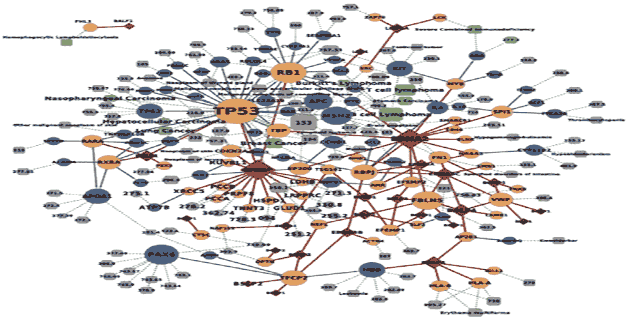 Viral Disease Networks, comprised of the viral neighborhoods in the host interactome, along with their disease associations, contain potential connections from viral targets to virally implicated diseases. Some of these connections are already known to be informative upon disease mechanisms, while others could be further candidates for investigations into the molecular mechanisms of diseases.
Viral Disease Networks, comprised of the viral neighborhoods in the host interactome, along with their disease associations, contain potential connections from viral targets to virally implicated diseases. Some of these connections are already known to be informative upon disease mechanisms, while others could be further candidates for investigations into the molecular mechanisms of diseases.
- Viral Disease Networks also contain diseases whose associations with viruses have not yet been recognized. We prioritized these diseases as candidate virally implicated diseases based on network topology, and benchmarked this prioritization of candidate diseases using population-based clinical associations between candidate diseases and viral infection.
- Experimental testing of several of these prioritizations uncovered evidence for a disease pathway that links HPV infection to Fanconi anemia.
The systematic viral disease-ome framework applied here acts to decipher the interplay between viruses and disease phenotypes, in line with the premise of this CEGS, that phenotypic effects of functional sequence variants are mediated through dynamic intracellular networks.