 Emerging exome and complete genome sequencing efforts have identified germline variations associated with cancer predisposition, and have catalogued thousands of somatic genomic alterations. Catalogs of cancer-associated genomic alterations require functional information to interpret their role in tumorigenesis. The challenge is to identify specific causal genes, given the enormous number of cancer-associated genomic variations these genomic approaches uncover.
Emerging exome and complete genome sequencing efforts have identified germline variations associated with cancer predisposition, and have catalogued thousands of somatic genomic alterations. Catalogs of cancer-associated genomic alterations require functional information to interpret their role in tumorigenesis. The challenge is to identify specific causal genes, given the enormous number of cancer-associated genomic variations these genomic approaches uncover.
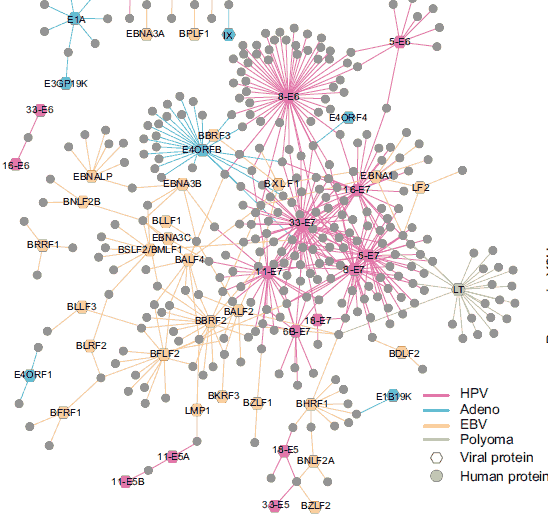
Using a systematic integrated pipeline investigated at genome-scale perturbations of host interactome and transcriptome networks induced by individual gene products encoded by members of four functionally related, yet biologically distinct, families of DNA tumor viruses (Rozenblatt-Rosen et al, Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins, Nature, 2012).
- Binary Interactome Network
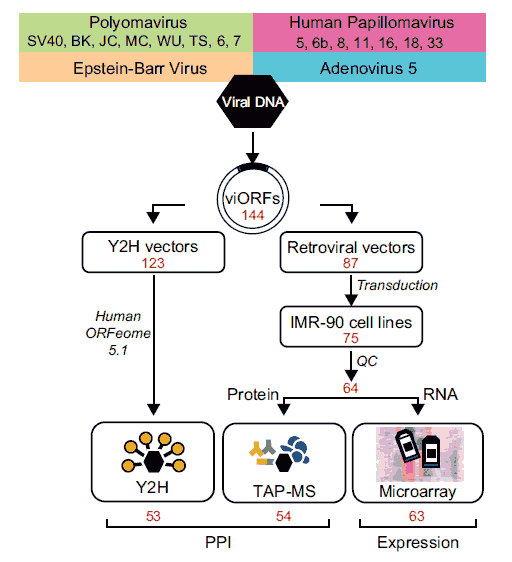
 To map binary physical interactions at genome-scale between viral and host proteins we used a stringent implementation of the yeast two-hybrid (Y2H) system, screening 123 viral ORFs against ~13,000 human ORFs. The viral-host binary interactome so obtained contains 454 validated binary interactions between 53 viral proteins and 307 human target proteins.
To map binary physical interactions at genome-scale between viral and host proteins we used a stringent implementation of the yeast two-hybrid (Y2H) system, screening 123 viral ORFs against ~13,000 human ORFs. The viral-host binary interactome so obtained contains 454 validated binary interactions between 53 viral proteins and 307 human target proteins.
- Co-complex Interactome Network
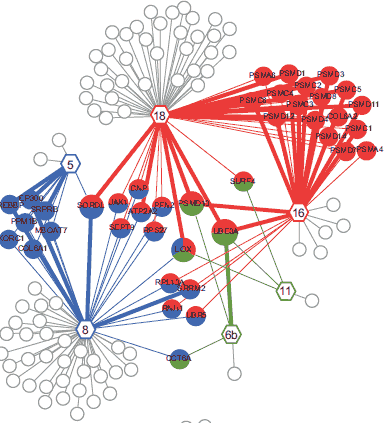 To map co-complex associations at proteome-scale between viral proteins and the host proteome, we implemented a tandem affinity purification protocol followed by mass spectrometry (TAP-MS). For TAP-MS we generated expression constructs of each viral ORF fused to a tandem epitope tag, and introduced each expression construct into IMR90 normal human diploid fibroblast cells. The intersection of two independent TAP-MS experiments yielded 3,787 reproducibly mapped viral-host co-complex associations involving 54 viral proteins and the products of 1,079 unambiguously identified host genes.
To map co-complex associations at proteome-scale between viral proteins and the host proteome, we implemented a tandem affinity purification protocol followed by mass spectrometry (TAP-MS). For TAP-MS we generated expression constructs of each viral ORF fused to a tandem epitope tag, and introduced each expression construct into IMR90 normal human diploid fibroblast cells. The intersection of two independent TAP-MS experiments yielded 3,787 reproducibly mapped viral-host co-complex associations involving 54 viral proteins and the products of 1,079 unambiguously identified host genes.
- Transcriptome Profiles
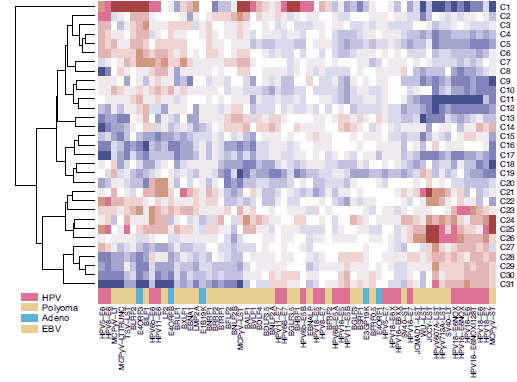 To characterize virally induced transcriptional perturbations at genome-scale, we completed microarray analyses on the TAP-tagged viral ORF-expressing IMR90 cell lines, scoring significant host gene expression changes. To identify patterns of host transcriptional perturbation common across the set of viral proteins, model-based clustering was used to construct clusters from the most frequently perturbed host genes. We identified 31 clusters of five or more genes, most of them enriched for GO terms and KEGG pathways.
To characterize virally induced transcriptional perturbations at genome-scale, we completed microarray analyses on the TAP-tagged viral ORF-expressing IMR90 cell lines, scoring significant host gene expression changes. To identify patterns of host transcriptional perturbation common across the set of viral proteins, model-based clustering was used to construct clusters from the most frequently perturbed host genes. We identified 31 clusters of five or more genes, most of them enriched for GO terms and KEGG pathways.
All the datasets accumulated from this pipeline are now publicly available (here).
Integrating together our three large-scale datasets of viral perturbations points to multiple pathways that go awry in cancer. A particular example is the Notch signaling pathway, which is significantly targeted by multiple viral proteins from all four of the viral groups tested in our genomics pipeline. 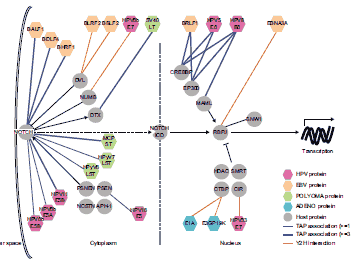 Aberrant Notch signaling can have either an oncogenic or tumor-suppressive role in human cancers, so the viral perturbations of Notch that we uncovered highlight hypotheses about how the Notch pathway might get derailed in disease. Experimentally following up on one of the hypotheses, we accumulated evidence that supports provisional observations implicating mutations in MAML1, a Notch pathway component, in pathogenesis.
Aberrant Notch signaling can have either an oncogenic or tumor-suppressive role in human cancers, so the viral perturbations of Notch that we uncovered highlight hypotheses about how the Notch pathway might get derailed in disease. Experimentally following up on one of the hypotheses, we accumulated evidence that supports provisional observations implicating mutations in MAML1, a Notch pathway component, in pathogenesis.
Integrating our pipeline datasets with sets of genes mutated in cancer showed that our systematic analysis of host targets of viral proteins identifies somatically mutated cancer genes with a success rate that is on par with and complementary to their identification by large-scale genome-wide efforts. The central hypothesis of this CEGS, that viral pathogens and human genetic variations similarly perturb properties of networks to induce disease, is thereby justified.