 Comparative interactomics, in contrast to standard sequence similarity metrics, measures the divergence of protein interaction profiles among closely related organisms (Kiemer & Cesareni, Trends Biotechnol 2007). The approach can correlate specific traits to phenotypic differences. We applied comparative interactomics at large scale to human papillomavirus (HPVs) E6 and E7 oncoproteins. We mapped the virus–host protein interaction networks of E6 and E7 proteins from 11 distinct HPV genotypes, selected for their different tropisms and pathologies.
Comparative interactomics, in contrast to standard sequence similarity metrics, measures the divergence of protein interaction profiles among closely related organisms (Kiemer & Cesareni, Trends Biotechnol 2007). The approach can correlate specific traits to phenotypic differences. We applied comparative interactomics at large scale to human papillomavirus (HPVs) E6 and E7 oncoproteins. We mapped the virus–host protein interaction networks of E6 and E7 proteins from 11 distinct HPV genotypes, selected for their different tropisms and pathologies.
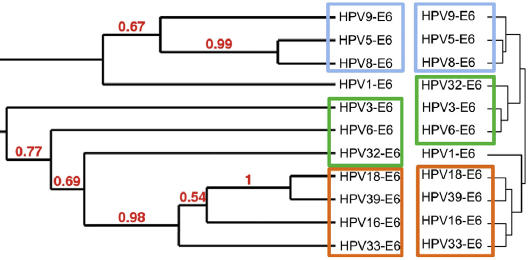
We combined two complementary binary protein interaction assays to produce robust and comprehensive virus-host interactome datasets: yeast two-hybrid (Y2H) and high-throughput Gaussia princeps protein complementation assay (HT-GPCA) [Cassonnet et al, Nat Methods 2011]. HT-GPCA detects protein interaction by measuring the interaction-mediated reconstitution of activity of a split G. princeps luciferase. Hierarchical clustering of interaction profiles of E6 (right of Figure) significantly recapitulated HPV phylogeny (left of Figure). Specific virus–host interaction profiles correlated with pathological traits and reflected the distinct carcinogenic potentials of different HPVs. Comparative interactomics accordingly constitutes a helpful strategy in line with the goals of this CEGS to decipher the molecular relationships underpinning pathogen–host interactions.