Genomic Analysis of Network Perturbations in Human Disease
Genetic differences between individuals influence susceptibility to disease. The Human Genome Project, together with the HapMap project and the Human Cancer Genome Project, have greatly accelerated the ability to find genetic variants and associated disease genes. Despite these advances, linking individual genes and their variations to disease remains daunting. Even where a causal variant has been identified, the biological insights have been slow in coming. 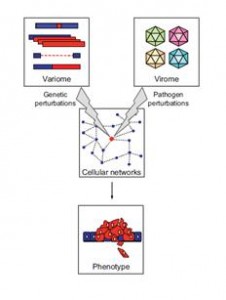 The phenotypic effects of functional sequence variants are mediated through dynamic networks of gene products and metabolites. Making sense of genotype-phenotype relationships thus requires that phenotypes be viewed as manifestations of network properties, rather than simply the result of genomic variations considered individually.
The phenotypic effects of functional sequence variants are mediated through dynamic networks of gene products and metabolites. Making sense of genotype-phenotype relationships thus requires that phenotypes be viewed as manifestations of network properties, rather than simply the result of genomic variations considered individually.
The central hypothesis of this CEGS is that both human genetic variations and pathogens such as viruses similarly influence local and global properties of networks to induce disease states. Our approach to understanding cellular networks is to observe perturbations of network structure by viral pathogen proteins, measuring the effects using interactome mapping, proteomic analysis, and transcriptional profiling.
We have been integrating measurements of network-level perturbations caused by four selected human DNA tumor virus families (polyomaviruses, particularly SV40, papillomaviruses, adenoviruses, and Epstein-Barr virus). The goal is to develop testable hypotheses that present insights into pathology of human disease.
Achievements
 Viral perturbations of host networks reflect disease etiology
Viral perturbations of host networks reflect disease etiology
Many virally implicated human diseases are associated with genetic alterations in particular disease susceptibility genes. For two human DNA tumor viruses, Epstein-Barr virus and human papillomavirus 16, we introduced a network-based framework to model the phenotypic consequences of the viral-host interactions. More…
 Global landscape of host perturbations by tumor virus proteins
Global landscape of host perturbations by tumor virus proteins
Emerging exome and complete genome sequencing efforts have identified germline variations associated with cancer predisposition, and have catalogued thousands of somatic genomic alterations. Catalogs of cancer-associated genomic alterations require functional information to interpret their consequences. More…
 FAM111A and SV40 large T antigen host range activity
FAM111A and SV40 large T antigen host range activity
Virus replication depends on interactions between viral and host cell proteins to overcome cellular defenses. The SV40 large T antigen (T-Ag) oncoprotein binds to numerous cellular proteins to enable efficient viral replication. More…
 Comparative interactomics of papillomavirus viral-host interactions
Comparative interactomics of papillomavirus viral-host interactions
Comparative interactomics, that is, homology by network comparison rather than sequence similarity, was applied at large scale to human papillomavirus (HPV) E6 and E7 proteins from 11 distinct HPV genotypes. More…